Browsing a HDF5 file.
HDF5 is a very general file format that permits to store data at various "semantic" or "data schema" defined by users. ioda knows two data schemas, the one of the now deprecated OpenScintist/BatchLab package and the one of g4tools/HDF5 introduced in the version 10.3 of Geant4.
For HDF5 and Geant4, see web pages at :
https://support.hdfgroup.org/HDF5
g4b4.hdf5
We are going to play with g4b4.hdf5 file produced by the B4 Geant4 novice example coming with the application under the resource directory.
Then start ioda and select the file with :
Files -> <resource> -> .hdf5 -> g4b4.hdf5
Whence selected you should see the list of the "H5 groups" representing directories (in grey) and plottable objects (in white). A HDF5 file can be organized in a hierarchy of H5groups with leaf "H5 datasets". It is the data schema/semantic that determines that a group must be understood as a directory or a container of leaf datasets defining an object as an histogram or an ntuple. At opening of the file, ioda analyses the groups and sees if he can find histos and ntuples.
The g4b4.hdf5 has two directories : default_hisgorams and default_ntuples. If choosing default_hisograms, you should a list with four histos : Eabs, Egap, Labs, Lgap. Choosing one of these permit to plot it. For example Egap leads to :
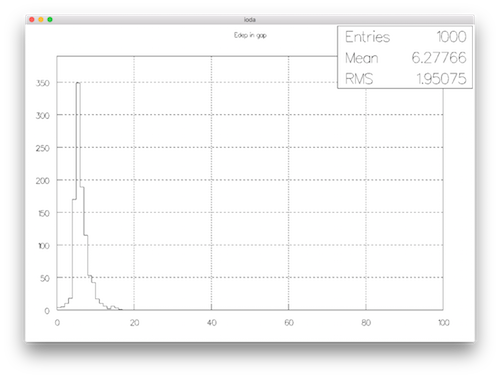
From here you can, for example, produce a JPEG file with :
meta-zone > Home > export > out.jpg
(meta-zone = click/touch the bottom area window that permits to toggle screen/menu mode).